-
Notifications
You must be signed in to change notification settings - Fork 2.5k
[HUDI-4363] Support Clustering row writer to improve performance #6046
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Changes from 10 commits
6bb8dd6
d94e3ec
25f1afd
fa7c100
1c91345
f9bed48
687ec04
b8e848d
5a16d35
60ef514
1355882
7300c9e
67924e5
090434e
988e487
f2bb9e6
1587f47
20f64af
9632919
2ff0b70
e75f6d0
46004b0
af01ca4
File filter
Filter by extension
Conversations
Jump to
Diff view
Diff view
There are no files selected for viewing
| Original file line number | Diff line number | Diff line change |
|---|---|---|
|
|
@@ -37,6 +37,7 @@ | |
| import org.apache.hudi.common.model.RewriteAvroPayload; | ||
| import org.apache.hudi.common.table.HoodieTableConfig; | ||
| import org.apache.hudi.common.table.log.HoodieMergedLogRecordScanner; | ||
| import org.apache.hudi.common.util.CollectionUtils; | ||
| import org.apache.hudi.common.util.FutureUtils; | ||
| import org.apache.hudi.common.util.Option; | ||
| import org.apache.hudi.common.util.StringUtils; | ||
|
|
@@ -47,8 +48,11 @@ | |
| import org.apache.hudi.exception.HoodieClusteringException; | ||
| import org.apache.hudi.exception.HoodieIOException; | ||
| import org.apache.hudi.execution.bulkinsert.BulkInsertInternalPartitionerFactory; | ||
| import org.apache.hudi.execution.bulkinsert.BulkInsertInternalPartitionerWithRowsFactory; | ||
| import org.apache.hudi.execution.bulkinsert.RDDCustomColumnsSortPartitioner; | ||
| import org.apache.hudi.execution.bulkinsert.RDDSpatialCurveSortPartitioner; | ||
| import org.apache.hudi.execution.bulkinsert.RowCustomColumnsSortPartitioner; | ||
| import org.apache.hudi.execution.bulkinsert.RowSpatialCurveSortPartitioner; | ||
| import org.apache.hudi.io.IOUtils; | ||
| import org.apache.hudi.io.storage.HoodieFileReader; | ||
| import org.apache.hudi.io.storage.HoodieFileReaderFactory; | ||
|
|
@@ -68,9 +72,13 @@ | |
| import org.apache.log4j.Logger; | ||
| import org.apache.spark.api.java.JavaRDD; | ||
| import org.apache.spark.api.java.JavaSparkContext; | ||
| import org.apache.spark.sql.Dataset; | ||
| import org.apache.spark.sql.Row; | ||
| import org.apache.spark.sql.SQLContext; | ||
|
|
||
| import java.io.IOException; | ||
| import java.util.ArrayList; | ||
| import java.util.Collection; | ||
| import java.util.Iterator; | ||
| import java.util.List; | ||
| import java.util.Map; | ||
|
|
@@ -98,10 +106,18 @@ public HoodieWriteMetadata<HoodieData<WriteStatus>> performClustering(final Hood | |
| // execute clustering for each group async and collect WriteStatus | ||
| Stream<HoodieData<WriteStatus>> writeStatusesStream = FutureUtils.allOf( | ||
| clusteringPlan.getInputGroups().stream() | ||
| .map(inputGroup -> runClusteringForGroupAsync(inputGroup, | ||
| clusteringPlan.getStrategy().getStrategyParams(), | ||
| Option.ofNullable(clusteringPlan.getPreserveHoodieMetadata()).orElse(false), | ||
| instantTime)) | ||
| .map(inputGroup -> { | ||
| if (getWriteConfig().getBooleanOrDefault("hoodie.datasource.write.row.writer.enable", false)) { | ||
|
Contributor
There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. Let's abstract this as a method in WriteConfig
Contributor
Author
There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more.
|
||
| return runClusteringForGroupAsyncWithRow(inputGroup, | ||
| clusteringPlan.getStrategy().getStrategyParams(), | ||
| Option.ofNullable(clusteringPlan.getPreserveHoodieMetadata()).orElse(false), | ||
|
||
| instantTime); | ||
| } | ||
| return runClusteringForGroupAsyncWithRDD(inputGroup, | ||
| clusteringPlan.getStrategy().getStrategyParams(), | ||
| Option.ofNullable(clusteringPlan.getPreserveHoodieMetadata()).orElse(false), | ||
| instantTime); | ||
| }) | ||
| .collect(Collectors.toList())) | ||
| .join() | ||
| .stream(); | ||
|
|
@@ -113,6 +129,15 @@ public HoodieWriteMetadata<HoodieData<WriteStatus>> performClustering(final Hood | |
| return writeMetadata; | ||
| } | ||
|
|
||
| /** | ||
| * Execute clustering to write inputRecords into new files based on strategyParams. | ||
| * Different from {@link performClusteringWithRecordsRDD}, this method take {@link Dataset<Row>} | ||
| * as inputs. | ||
| */ | ||
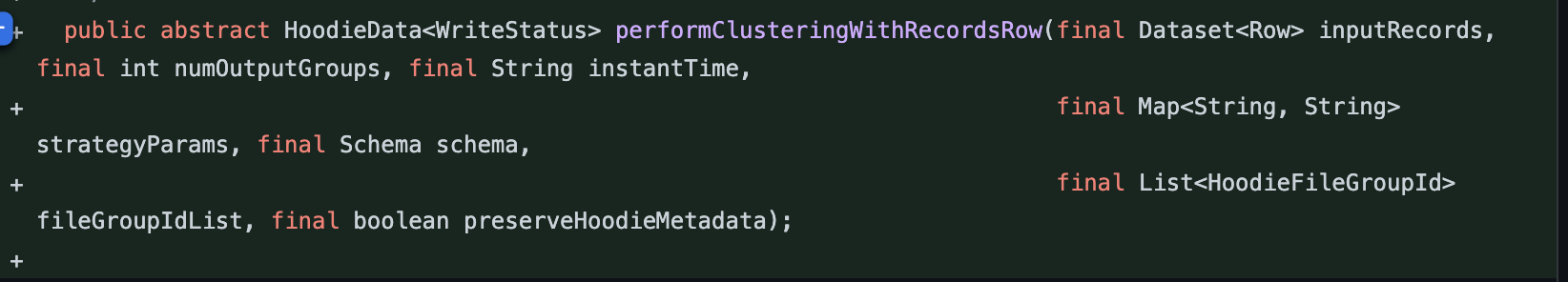
| public abstract HoodieData<WriteStatus> performClusteringWithRecordsRow(final Dataset<Row> inputRecords, final int numOutputGroups, final String instantTime, | ||
|
||
| final Map<String, String> strategyParams, final Schema schema, | ||
| final List<HoodieFileGroupId> fileGroupIdList, final boolean preserveHoodieMetadata); | ||
|
|
||
| /** | ||
| * Execute clustering to write inputRecords into new files as defined by rules in strategy parameters. | ||
| * The number of new file groups created is bounded by numOutputGroups. | ||
|
|
@@ -131,14 +156,25 @@ public abstract HoodieData<WriteStatus> performClusteringWithRecordsRDD(final Ho | |
| final Map<String, String> strategyParams, final Schema schema, | ||
| final List<HoodieFileGroupId> fileGroupIdList, final boolean preserveHoodieMetadata); | ||
|
|
||
| protected BulkInsertPartitioner<Dataset<Row>> getRowPartitioner(Map<String, String> strategyParams, | ||
| Schema schema) { | ||
| return getPartitioner(strategyParams, schema, true); | ||
| } | ||
|
|
||
| protected BulkInsertPartitioner<JavaRDD<HoodieRecord<T>>> getRDDPartitioner(Map<String, String> strategyParams, | ||
| Schema schema) { | ||
| return getPartitioner(strategyParams, schema, false); | ||
| } | ||
|
|
||
| /** | ||
| * Create {@link BulkInsertPartitioner} based on strategy params. | ||
| * | ||
| * @param strategyParams Strategy parameters containing columns to sort the data by when clustering. | ||
| * @param schema Schema of the data including metadata fields. | ||
| * @return {@link RDDCustomColumnsSortPartitioner} if sort columns are provided, otherwise empty. | ||
| */ | ||
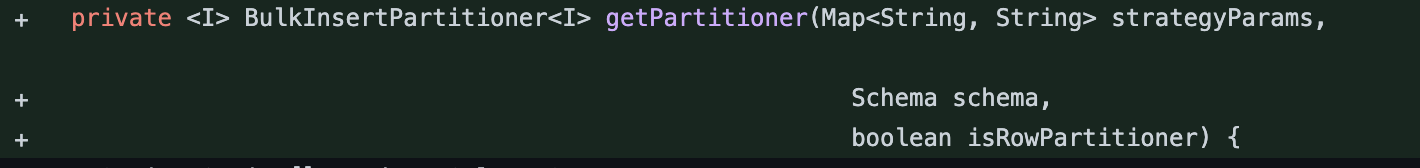
| protected BulkInsertPartitioner<JavaRDD<HoodieRecord<T>>> getPartitioner(Map<String, String> strategyParams, Schema schema) { | ||
| private <I> BulkInsertPartitioner<I> getPartitioner(Map<String, String> strategyParams, | ||
| Schema schema, | ||
| boolean isRowPartitioner) { | ||
| Option<String[]> orderByColumnsOpt = | ||
| Option.ofNullable(strategyParams.get(PLAN_STRATEGY_SORT_COLUMNS.key())) | ||
| .map(listStr -> listStr.split(",")); | ||
|
|
@@ -148,29 +184,34 @@ protected BulkInsertPartitioner<JavaRDD<HoodieRecord<T>>> getPartitioner(Map<Str | |
| switch (layoutOptStrategy) { | ||
| case ZORDER: | ||
| case HILBERT: | ||
| return new RDDSpatialCurveSortPartitioner( | ||
| return isRowPartitioner | ||
| ? new RowSpatialCurveSortPartitioner(getWriteConfig()) | ||
| : new RDDSpatialCurveSortPartitioner( | ||
| (HoodieSparkEngineContext) getEngineContext(), | ||
|
||
| orderByColumns, | ||
| layoutOptStrategy, | ||
| getWriteConfig().getLayoutOptimizationCurveBuildMethod(), | ||
| HoodieAvroUtils.addMetadataFields(schema)); | ||
| case LINEAR: | ||
| return new RDDCustomColumnsSortPartitioner(orderByColumns, HoodieAvroUtils.addMetadataFields(schema), | ||
| return isRowPartitioner | ||
| ? new RowCustomColumnsSortPartitioner(orderByColumns) | ||
| : new RDDCustomColumnsSortPartitioner(orderByColumns, HoodieAvroUtils.addMetadataFields(schema), | ||
| getWriteConfig().isConsistentLogicalTimestampEnabled()); | ||
|
||
| default: | ||
| throw new UnsupportedOperationException(String.format("Layout optimization strategy '%s' is not supported", layoutOptStrategy)); | ||
| } | ||
| }).orElse(BulkInsertInternalPartitionerFactory.get(getWriteConfig().getBulkInsertSortMode())); | ||
| }).orElse(isRowPartitioner ? BulkInsertInternalPartitionerWithRowsFactory.get(getWriteConfig().getBulkInsertSortMode()) : | ||
| BulkInsertInternalPartitionerFactory.get(getWriteConfig().getBulkInsertSortMode())); | ||
| } | ||
|
|
||
| /** | ||
| * Submit job to execute clustering for the group. | ||
| * Submit job to execute clustering for the group with RDD APIs. | ||
| */ | ||
| private CompletableFuture<HoodieData<WriteStatus>> runClusteringForGroupAsync(HoodieClusteringGroup clusteringGroup, Map<String, String> strategyParams, | ||
| boolean preserveHoodieMetadata, String instantTime) { | ||
| private CompletableFuture<HoodieData<WriteStatus>> runClusteringForGroupAsyncWithRDD(HoodieClusteringGroup clusteringGroup, Map<String, String> strategyParams, | ||
|
||
| boolean preserveHoodieMetadata, String instantTime) { | ||
| return CompletableFuture.supplyAsync(() -> { | ||
| JavaSparkContext jsc = HoodieSparkEngineContext.getSparkContext(getEngineContext()); | ||
| HoodieData<HoodieRecord<T>> inputRecords = readRecordsForGroup(jsc, clusteringGroup, instantTime); | ||
| HoodieData<HoodieRecord<T>> inputRecords = readRecordsForGroupAsRDD(jsc, clusteringGroup, instantTime); | ||
| Schema readerSchema = HoodieAvroUtils.addMetadataFields(new Schema.Parser().parse(getWriteConfig().getSchema())); | ||
| List<HoodieFileGroupId> inputFileIds = clusteringGroup.getSlices().stream() | ||
| .map(info -> new HoodieFileGroupId(info.getPartitionPath(), info.getFileId())) | ||
|
|
@@ -179,10 +220,26 @@ private CompletableFuture<HoodieData<WriteStatus>> runClusteringForGroupAsync(Ho | |
| }); | ||
| } | ||
|
|
||
| /** | ||
| * Submit job to execute clustering for the group with dataset APIs. | ||
| */ | ||
| private CompletableFuture<HoodieData<WriteStatus>> runClusteringForGroupAsyncWithRow(HoodieClusteringGroup clusteringGroup, Map<String, String> strategyParams, | ||
| boolean preserveHoodieMetadata, String instantTime) { | ||
| return CompletableFuture.supplyAsync(() -> { | ||
| JavaSparkContext jsc = HoodieSparkEngineContext.getSparkContext(getEngineContext()); | ||
| Dataset<Row> inputRecords = readRecordsForGroupAsRow(jsc, clusteringGroup, instantTime); | ||
| Schema readerSchema = HoodieAvroUtils.addMetadataFields(new Schema.Parser().parse(getWriteConfig().getSchema())); | ||
| List<HoodieFileGroupId> inputFileIds = clusteringGroup.getSlices().stream() | ||
| .map(info -> new HoodieFileGroupId(info.getPartitionPath(), info.getFileId())) | ||
| .collect(Collectors.toList()); | ||
| return performClusteringWithRecordsRow(inputRecords, clusteringGroup.getNumOutputFileGroups(), instantTime, strategyParams, readerSchema, inputFileIds, preserveHoodieMetadata); | ||
| }); | ||
| } | ||
|
|
||
| /** | ||
| * Get RDD of all records for the group. This includes all records from file slice (Apply updates from log files, if any). | ||
| */ | ||
| private HoodieData<HoodieRecord<T>> readRecordsForGroup(JavaSparkContext jsc, HoodieClusteringGroup clusteringGroup, String instantTime) { | ||
| private HoodieData<HoodieRecord<T>> readRecordsForGroupAsRDD(JavaSparkContext jsc, HoodieClusteringGroup clusteringGroup, String instantTime) { | ||
| List<ClusteringOperation> clusteringOps = clusteringGroup.getSlices().stream().map(ClusteringOperation::create).collect(Collectors.toList()); | ||
| boolean hasLogFiles = clusteringOps.stream().anyMatch(op -> op.getDeltaFilePaths().size() > 0); | ||
| if (hasLogFiles) { | ||
|
|
@@ -273,6 +330,60 @@ private HoodieData<HoodieRecord<T>> readRecordsForGroupBaseFiles(JavaSparkContex | |
| .map(record -> transform(record, writeConfig))); | ||
| } | ||
|
|
||
| /** | ||
| * Get dataset of all records for the group. This includes all records from file slice (Apply updates from log files, if any). | ||
| */ | ||
| private Dataset<Row> readRecordsForGroupAsRow(JavaSparkContext jsc, | ||
| HoodieClusteringGroup clusteringGroup, | ||
| String instantTime) { | ||
| List<ClusteringOperation> clusteringOps = clusteringGroup.getSlices().stream() | ||
| .map(ClusteringOperation::create).collect(Collectors.toList()); | ||
| boolean hasLogFiles = clusteringOps.stream().anyMatch(op -> op.getDeltaFilePaths().size() > 0); | ||
| SQLContext sqlContext = new SQLContext(jsc.sc()); | ||
|
|
||
| String[] baseFilePaths = clusteringOps | ||
| .stream() | ||
| .map(op -> { | ||
| ArrayList<String> readPaths = new ArrayList<>(); | ||
| if (op.getBootstrapFilePath() != null) { | ||
| readPaths.add(op.getBootstrapFilePath()); | ||
| } | ||
| if (op.getDataFilePath() != null) { | ||
| readPaths.add(op.getDataFilePath()); | ||
| } | ||
| return readPaths; | ||
| }) | ||
| .flatMap(Collection::stream) | ||
| .filter(path -> !path.isEmpty()) | ||
| .toArray(String[]::new); | ||
| String[] deltaPaths = clusteringOps | ||
| .stream() | ||
| .filter(op -> !op.getDeltaFilePaths().isEmpty()) | ||
| .flatMap(op -> op.getDeltaFilePaths().stream()) | ||
| .toArray(String[]::new); | ||
|
|
||
| Dataset<Row> inputRecords; | ||
| if (hasLogFiles) { | ||
| String compactionFractor = Option.ofNullable(getWriteConfig().getString("compaction.memory.fraction")) | ||
|
Contributor
There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. The only part that differs b/w these branches are the options composition. Let's extract the common part and only keep options composition under conditional
Contributor
Author
There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. done |
||
| .orElse("0.75"); | ||
| String[] paths = CollectionUtils.combine(baseFilePaths, deltaPaths); | ||
| inputRecords = sqlContext.read() | ||
|
||
| .format("org.apache.hudi") | ||
| .option("hoodie.datasource.query.type", "snapshot") | ||
| .option("compaction.memory.fraction", compactionFractor) | ||
| .option("as.of.instant", instantTime) | ||
| .option("hoodie.datasource.read.paths", String.join(",", paths)) | ||
| .load(); | ||
| } else { | ||
| inputRecords = sqlContext.read() | ||
| .format("org.apache.hudi") | ||
| .option("as.of.instant", instantTime) | ||
|
||
| .option("hoodie.datasource.read.paths", String.join(",", baseFilePaths)) | ||
| .load(); | ||
| } | ||
| return inputRecords; | ||
| } | ||
|
|
||
| /** | ||
| * Stream to array conversion with generic type is not straightforward. | ||
| * Implement a utility method to abstract high level logic. This needs to be improved in future | ||
|
|
||
| Original file line number | Diff line number | Diff line change |
|---|---|---|
|
|
@@ -33,8 +33,11 @@ | |
| import org.apache.hudi.table.action.commit.SparkBulkInsertHelper; | ||
|
|
||
| import org.apache.avro.Schema; | ||
| import org.apache.hudi.table.action.commit.SparkBulkInsertRowWriter; | ||
| import org.apache.log4j.LogManager; | ||
| import org.apache.log4j.Logger; | ||
| import org.apache.spark.sql.Dataset; | ||
| import org.apache.spark.sql.Row; | ||
|
|
||
| import java.util.List; | ||
| import java.util.Map; | ||
|
|
@@ -54,6 +57,35 @@ public SparkSingleFileSortExecutionStrategy(HoodieTable table, | |
| super(table, engineContext, writeConfig); | ||
| } | ||
|
|
||
| @Override | ||
| public HoodieData<WriteStatus> performClusteringWithRecordsRow(Dataset<Row> inputRecords, | ||
| int numOutputGroups, | ||
| String instantTime, | ||
| Map<String, String> strategyParams, | ||
| Schema schema, | ||
| List<HoodieFileGroupId> fileGroupIdList, | ||
| boolean preserveHoodieMetadata) { | ||
| if (numOutputGroups != 1 || fileGroupIdList.size() != 1) { | ||
| throw new HoodieClusteringException("Expect only one file group for strategy: " + getClass().getName()); | ||
| } | ||
| LOG.info("Starting clustering for a group, parallelism:" + numOutputGroups + " commit:" + instantTime); | ||
|
|
||
| HoodieWriteConfig newConfig = HoodieWriteConfig.newBuilder() | ||
| .withBulkInsertParallelism(numOutputGroups) | ||
| .withProps(getWriteConfig().getProps()).build(); | ||
|
|
||
| boolean shouldPreserveHoodieMetadata = preserveHoodieMetadata; | ||
| if (!newConfig.populateMetaFields() && preserveHoodieMetadata) { | ||
| LOG.warn("Will setting preserveHoodieMetadata to false as populateMetaFields is false"); | ||
| shouldPreserveHoodieMetadata = false; | ||
| } | ||
|
|
||
| newConfig.setValue(HoodieStorageConfig.PARQUET_MAX_FILE_SIZE, String.valueOf(Long.MAX_VALUE)); | ||
|
Contributor
There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. Let's duplicate the comment from the original method as well |
||
|
|
||
| return SparkBulkInsertRowWriter.bulkInsert(inputRecords, instantTime, getHoodieTable(), newConfig, | ||
| getRowPartitioner(strategyParams, schema), numOutputGroups, shouldPreserveHoodieMetadata); | ||
| } | ||
|
|
||
| @Override | ||
| public HoodieData<WriteStatus> performClusteringWithRecordsRDD(HoodieData<HoodieRecord<T>> inputRecords, | ||
| int numOutputGroups, | ||
|
|
@@ -74,6 +106,6 @@ public HoodieData<WriteStatus> performClusteringWithRecordsRDD(HoodieData<Hoodie | |
| newConfig.setValue(HoodieStorageConfig.PARQUET_MAX_FILE_SIZE, String.valueOf(Long.MAX_VALUE)); | ||
|
|
||
| return (HoodieData<WriteStatus>) SparkBulkInsertHelper.newInstance().bulkInsert(inputRecords, instantTime, getHoodieTable(), newConfig, | ||
| false, getPartitioner(strategyParams, schema), true, numOutputGroups, new SingleFileHandleCreateFactory(fileGroupIdList.get(0).getFileId(), preserveHoodieMetadata)); | ||
| false, getRDDPartitioner(strategyParams, schema), true, numOutputGroups, new SingleFileHandleCreateFactory(fileGroupIdList.get(0).getFileId(), preserveHoodieMetadata)); | ||
| } | ||
| } | ||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
We need to do
getBooleanOrDefault, otherwise it might NPE (due to unboxing)There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
EMBEDDED_TIMELINE_SERVER_REUSE_ENABLEDis a ConfigProperty, getBoolean will handle this.