High-performance VCF to PostgreSQL loader with clinical-grade compliance, purpose-built for Polygenic Risk Score (PRS) research.
vcf-pg-loader provides a complete data infrastructure for polygenic risk score research:
| Feature | Description |
|---|---|
| GWAS Summary Statistics | Import and query GWAS results in GWAS-SSF standard format |
| PGS Catalog Integration | Load PRS weights directly from PGS Catalog scoring files |
| HapMap3 Reference Panel | Built-in support for HapMap3 SNPs used by PRS-CS, LDpred2, and other methods |
| LD Block Annotations | Berisa & Pickrell (2016) LD blocks for Bayesian PRS methods |
| Multi-Ancestry Frequencies | Population-specific allele frequencies from gnomAD for ancestry-aware PRS |
| Genotype Dosages | Imputation dosages and genotype probabilities (GP) for accurate PRS calculation |
| Sample QC Metrics | Call rate, het/hom ratio, Ti/Tv, sex inference, and contamination checks |
| Variant QC Metrics | HWE exact test, INFO score, call rate, MAF computed at load time |
| Materialized Views | Pre-computed PRS-ready variant sets with concurrent refresh |
| Export to PRS Tools | Direct export to PLINK, PRS-CS, LDpred2, and PRSice-2 formats |
- Streaming VCF parsing with cyvcf2 for memory-efficient processing
- Variant normalization using the vt algorithm (left-align and trim)
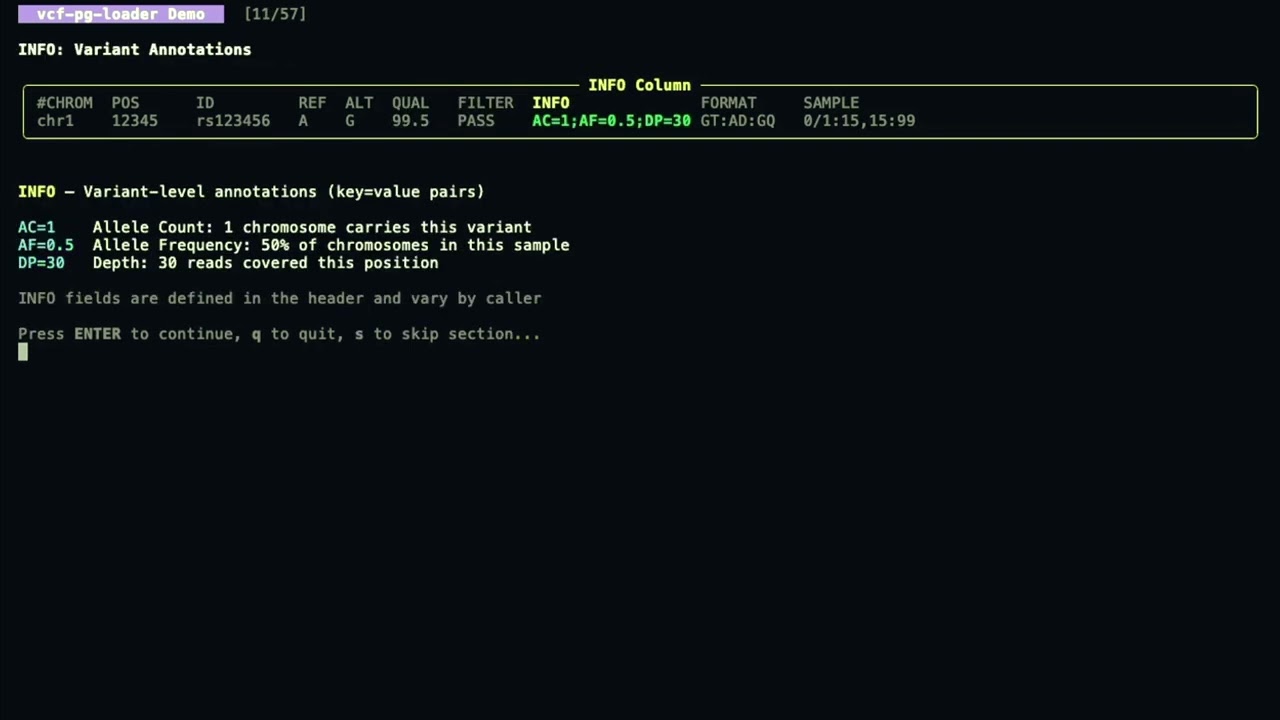
- Number=A/R/G field handling - proper per-ALT extraction during multi-allelic decomposition
- Binary COPY protocol via asyncpg for maximum insert performance
- Chromosome-partitioned tables for efficient region queries
- Human and non-human genome support - chromosome enum for human, TEXT for others
- GWAS summary statistics - import GWAS-SSF format files with study metadata
- PGS Catalog weights - load scoring files with automatic variant matching
- Reference panels - HapMap3 SNP sets for LD-aware PRS methods
- LD block definitions - genome partitioning for PRS-CS and SBayesR
- Population frequencies - multi-ancestry AF from gnomAD, 1000 Genomes
- Genotype storage - hash-partitioned with dosage and GP support
- Variant QC - HWE p-value, INFO score, call rate, AAF/MAF/MAC
- Sample QC - call rate, het/hom ratio, Ti/Tv, F coefficient, sex inference
- Materialized views - pre-filtered PRS candidate variants
- SQL functions - HWE exact test, allele harmonization in-database
- Audit trail with load batch tracking and validation
- CLI interface with Typer for easy operation
- TOML configuration - file-based configuration with CLI overrides
- Progress reporting - real-time progress bar with
rich - Docker support - multi-stage Dockerfile and docker-compose for development
- Zero-config database - auto-managed PostgreSQL via Docker, no setup required
conda install -c conda-forge -c bioconda vcf-pg-loaderpip install vcf-pg-loadercurl -fsSL https://raw.githubusercontent.com/Zacharyr41/vcf-pg-loader/main/install.sh | bashThis installs vcf-pg-loader and all dependencies (Python, Docker) automatically.
git clone https://github.com/Zacharyr41/vcf-pg-loader.git
cd vcf-pg-loader
uv pip install -e ".[dev]"The vcfpgloader/load module is available in nf-core/modules for use in Nextflow pipelines.
nf-core modules install vcfpgloader/loadinclude { VCFPGLOADER_LOAD } from '../modules/nf-core/vcfpgloader/load/main'
workflow {
ch_input = Channel.of([
[ id: 'sample1', family: 'FAM001' ], // meta map
file('sample1.vcf.gz'), // vcf
file('sample1.vcf.gz.tbi'), // tbi
'localhost', // db_host
5432, // db_port
'variants_db', // db_name
'postgres', // db_user
'public' // db_schema
])
VCFPGLOADER_LOAD(ch_input)
// Access outputs
VCFPGLOADER_LOAD.out.report // JSON report with loading statistics
VCFPGLOADER_LOAD.out.log // Detailed loading log
VCFPGLOADER_LOAD.out.row_count // Number of variants loaded
}Set PGPASSWORD via environment variable or Nextflow secrets:
// nextflow.config - Option 1: Environment variable
env {
PGPASSWORD = System.getenv('PGPASSWORD')
}
// nextflow.config - Option 2: Nextflow secrets
env {
PGPASSWORD = secrets.PGPASSWORD
}Customize batch size and other options via ext directives:
// nextflow.config
process {
withName: 'VCFPGLOADER_LOAD' {
ext.batch_size = '50000' // variants per batch (default: 10000)
ext.args = '--normalize' // additional CLI arguments
}
}| Channel | Description |
|---|---|
report |
JSON file with loading statistics (variants loaded, elapsed time, throughput) |
log |
Detailed loading log with warnings/errors |
row_count |
Integer count of variants successfully loaded |
versions |
Tool version for MultiQC reporting |
vcf-pg-loader doctorNo PostgreSQL setup required - vcf-pg-loader manages a local database automatically:
# Load a VCF file (auto-starts PostgreSQL in Docker)
vcf-pg-loader load sample.vcf.gz
# Check database status
vcf-pg-loader db status
# Open psql shell to query data
vcf-pg-loader db shell# Initialize database schema
vcf-pg-loader init-db --db postgresql://user:pass@localhost/variants
# Load a VCF file
vcf-pg-loader load sample.vcf.gz --db postgresql://user:pass@localhost/variants
# Validate a completed load
vcf-pg-loader validate <load-batch-id> --db postgresql://user:pass@localhost/variants# Load without normalization
vcf-pg-loader load sample.vcf.gz --no-normalize
# Load non-human VCF (e.g., SARS-CoV-2)
vcf-pg-loader load sarscov2.vcf.gz --no-human-genome
# Initialize for non-human genomes
vcf-pg-loader init-db --db postgresql://... --no-human-genome# 1. Load imputed VCF with genotype dosages
vcf-pg-loader load imputed.vcf.gz --db postgresql://localhost/prs_db
# 2. Import GWAS summary statistics
vcf-pg-loader import-gwas gwas_sumstats.tsv \
--study-id GCST90012345 \
--trait "Type 2 Diabetes" \
--db postgresql://localhost/prs_db
# 3. Load PGS Catalog weights
vcf-pg-loader import-pgs PGS000001_hmPOS_GRCh38.txt \
--db postgresql://localhost/prs_db
# 4. Load HapMap3 reference panel
vcf-pg-loader load-reference hapmap3.tsv \
--panel-name hapmap3 \
--db postgresql://localhost/prs_db
# 5. Annotate variants with LD blocks
vcf-pg-loader annotate-ld-blocks \
--population EUR \
--db postgresql://localhost/prs_db
# 6. Compute sample QC metrics
vcf-pg-loader compute-sample-qc \
--db postgresql://localhost/prs_db
# 7. Refresh materialized views for fast queries
vcf-pg-loader refresh-views --db postgresql://localhost/prs_db
# 8. Export to PRS-CS format
vcf-pg-loader export-prs-cs \
--study-id 1 \
--output gwas_prscs.txt \
--hapmap3-only \
--db postgresql://localhost/prs_dbLoad a VCF file into PostgreSQL.
vcf-pg-loader load <vcf_path> [OPTIONS]
Options:
--db, -d PostgreSQL connection URL (omit for auto-managed DB)
--batch, -b Records per batch [default: 50000]
--workers, -w Parallel workers [default: 8]
--normalize/--no-normalize Normalize variants using vt algorithm [default: normalize]
--drop-indexes/--keep-indexes Drop indexes during load [default: drop-indexes]
--human-genome/--no-human-genome Use human chromosome enum type [default: human-genome]
--config, -c TOML configuration file
--verbose, -v Enable verbose logging (DEBUG level)
--quiet, -q Suppress non-error output
--progress/--no-progress Show progress bar [default: progress]
--force, -f Force reload even if file was already loaded
--hipaa-mode/--no-hipaa-mode Enable/disable HIPAA compliance features [default: enabled]When --db is omitted, vcf-pg-loader automatically uses a managed PostgreSQL container.
Normalization: When enabled (default), variants are left-aligned and trimmed following the vt algorithm. This ensures consistent representation across different variant callers.
Genome Type: Human genome mode uses a PostgreSQL enum for chromosomes (chr1-22, X, Y, M) which provides validation and efficient storage. Non-human mode uses TEXT to support arbitrary chromosome/contig names.
HIPAA Mode: By default, vcf-pg-loader runs with HIPAA compliance features enabled:
- TLS required for database connections
- Sample ID anonymization
- VCF header sanitization to remove PHI
For local development without PHI, use --no-hipaa-mode to disable all compliance features:
vcf-pg-loader load sample.vcf.gz --no-hipaa-modeValidate a completed load by checking record counts and duplicates.
vcf-pg-loader validate <load_batch_id> [OPTIONS]
Options:
--db, -d PostgreSQL connection URLInitialize the database schema (tables, indexes, extensions).
vcf-pg-loader init-db [OPTIONS]
Options:
--db, -d PostgreSQL connection URL
--human-genome/--no-human-genome Use human chromosome enum type [default: human-genome]Important: The genome type must match between init-db and load commands. Use --no-human-genome for both when loading non-human VCFs.
Run performance benchmarks on VCF parsing and loading.
vcf-pg-loader benchmark [OPTIONS]
Options:
--vcf, -f Path to VCF file (uses built-in fixture if omitted)
--synthetic, -s Generate synthetic VCF with N variants
--db, -d PostgreSQL URL (omit for parsing-only benchmark)
--batch, -b Batch size [default: 50000]
--normalize/--no-normalize Test with/without normalization
--json Output results as JSON (for CI integration)
--quiet, -q Minimal outputExamples:
# Quick benchmark with built-in fixture (~2.6K variants)
vcf-pg-loader benchmark
# Generate and benchmark 100K synthetic variants
vcf-pg-loader benchmark --synthetic 100000
# Benchmark a specific VCF file
vcf-pg-loader benchmark --vcf /path/to/sample.vcf.gz
# Full benchmark including database loading
vcf-pg-loader benchmark --synthetic 50000 --db postgresql://localhost/variants
# JSON output for CI/scripting
vcf-pg-loader benchmark --synthetic 10000 --jsonSample output:
Benchmark Results (synthetic)
Variants: 100,000
Batch size: 50,000
Normalized: True
Parsing: 100,000 variants in 0.94s (106,000/sec)
Check system dependencies and diagnose issues.
vcf-pg-loader doctor
# Example output:
Dependency Check
Python 3.12.4 OK
cyvcf2 0.30.22 OK
asyncpg 0.29.0 OK
Docker 24.0.5 OK
Docker daemon running OKManage the local PostgreSQL database (Docker-based).
vcf-pg-loader db start # Start PostgreSQL container
vcf-pg-loader db stop # Stop the container
vcf-pg-loader db status # Show running status and connection URL
vcf-pg-loader db url # Print connection URL (for scripts)
vcf-pg-loader db shell # Open psql shell
vcf-pg-loader db reset # Remove container and all data- VCFHeaderParser - Parses VCF headers via cyvcf2's native API to extract INFO/FORMAT field definitions
- VCFStreamingParser - Memory-efficient streaming iterator that yields batches of
VariantRecordobjects - VariantParser - Handles per-variant parsing with Number=A/R/G field extraction for multi-allelic decomposition
- VCFLoader - Orchestrates loading with asyncpg binary COPY protocol
- SchemaManager - Manages PostgreSQL schema creation and index management
VCF File → VCFStreamingParser → Batch Buffer → asyncpg COPY → PostgreSQL
↓
VCFHeaderParser (field metadata)
↓
VariantParser (Number=A/R/G extraction)
This project was inspired by and builds upon several foundational tools in the genomics community:
Slivar - Rapid variant filtering:
Pedersen, B.S., Brown, J.M., Dashnow, H. et al. Effective variant filtering and expected candidate variant yield in studies of rare human disease. npj Genom. Med. 6, 60 (2021). https://doi.org/10.1038/s41525-021-00227-3
GEMINI - Original SQL-based VCF database:
Paila, U., Chapman, B.A., Kirchner, R., & Quinlan, A.R. GEMINI: Integrative Exploration of Genetic Variation and Genome Annotations. PLoS Comput Biol 9(7): e1003153 (2013). https://doi.org/10.1371/journal.pcbi.1003153
cyvcf2 - Python VCF parsing:
Pedersen, B.S. & Quinlan, A.R. cyvcf2: fast, flexible variant analysis with Python. Bioinformatics 33(12), 1867–1869 (2017). https://doi.org/10.1093/bioinformatics/btx057
- vcf2db: https://github.com/quinlan-lab/vcf2db
- VCF Format: Danecek et al. (2011) https://doi.org/10.1093/bioinformatics/btr330
- bcftools/HTSlib: Danecek et al. (2021) https://doi.org/10.1093/gigascience/giab008
- GIAB Benchmarks: Zook et al. (2019) https://doi.org/10.1038/s41587-019-0074-6
vcf-pg-loader supports TOML configuration files for persistent settings:
# vcf-pg-loader.toml
[vcf_pg_loader]
batch_size = 25000
workers = 16
normalize = true
drop_indexes = true
human_genome = true
log_level = "INFO"Use with the --config flag:
vcf-pg-loader load sample.vcf.gz --config vcf-pg-loader.tomlCLI arguments override config file values.
# Start PostgreSQL and run a load
docker-compose up -d postgres
docker-compose run vcf-pg-loader load /data/sample.vcf.gz --db postgresql://vcfloader:vcfloader@postgres:5432/variants
# Or build and run standalone
docker build -t vcf-pg-loader .
docker run vcf-pg-loader --helppostgres: PostgreSQL 16 with health checksvcf-pg-loader: The loader application
Mount your VCF files to /data in the container.
# Run all tests
uv run pytest
# Run with coverage
uv run pytest --cov=vcf_pg_loader
# Run only unit tests (skip integration)
uv run pytest -m "not integration"# Lint
uv run ruff check src tests
# Type check
uv run mypy src- CLI Reference - Complete command-line documentation
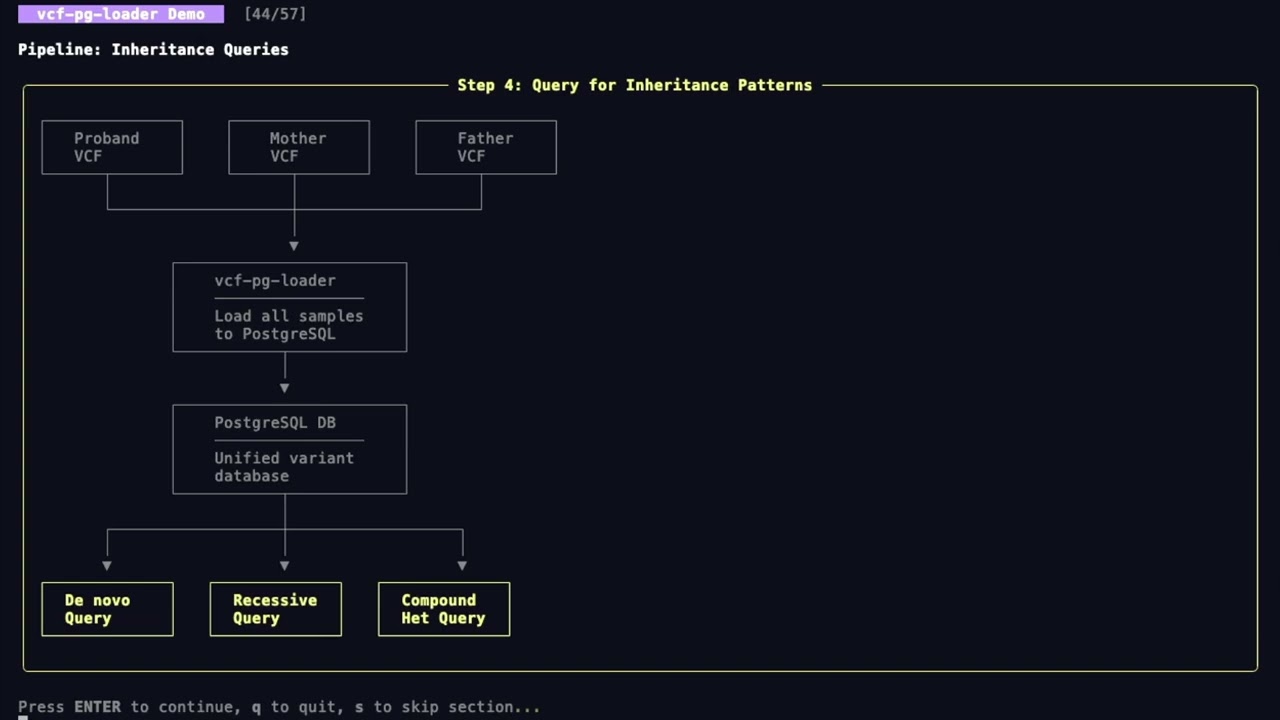
- PRS Workflows - End-to-end PRS analysis pipelines
- Schema Overview - Complete database schema with ER diagrams
- PRS Tables - PGS scores and weights storage
- GWAS Tables - Summary statistics (GWAS-SSF)
- Reference Tables - HapMap3, LD blocks
- Genotypes Tables - Individual-level data
- QC Tables - Sample and variant QC metrics
- Materialized Views - Pre-computed PRS query results
- Genomics Concepts - Understanding VCF data for non-geneticists
- Glossary of Terms - Technical terminology reference
- Architecture - Detailed system design and implementation
MIT - See LICENSE for details.